Thermococcus Eurythermalis Endonuclease IV Can Cleave Various Apurinic/Apyrimidinic Site Analogues in ssDNA and dsDNA
Endonuclease IV (EndoIV) is a DNA damage-specific endonuclease that mainly hydrolyzes the phosphodiester bond located at 5 of an apurinic/apyrimidinic (AP) site in DNA. EndoIV also possesses 3-exonuclease activity for removing 3-blocking groups and normal nucleotides. Here, we report that Thermococcus eurythermalis EndoIV (TeuendoIV) shows AP endonuclease and 3-exonuclease activities. The effect of AP site structures, positions and clustered patterns on the activity was characterized. The AP endonuclease activity of TeuendoIV can incise DNA 5 to various AP site analogues, including the alkane chain Spacer and polyethylene glycol Spacer. However, the short Spacer C2 strongly inhibits the AP endonuclease activity. The kinetic parameters also support its preference to various AP site analogues. In addition, the efficient cleavage at AP sites requires ≥2 normal nucleotides existing at the 5-terminus. The 3-exonuclease activity of TeuendoIV can remove one or more consecutive AP sites at the 3-terminus. Mutations on the residues for substrate recognition show that binding AP site-containing or complementary strand plays a key role for the hydrolysis of phosphodiester bonds. Our results provide a comprehensive biochemical characterization of the cleavage/removal of AP site analogues and some insight for repairing AP sites in hyperthermophile cells.
(Author: Wei-Wei Wang, Huan Zhou, Juan-Juan Xie, Gang-Shun Yi, Jian-Hua He, Feng-Ping Wang, Xiang Xiao and Xi-Peng Liu)
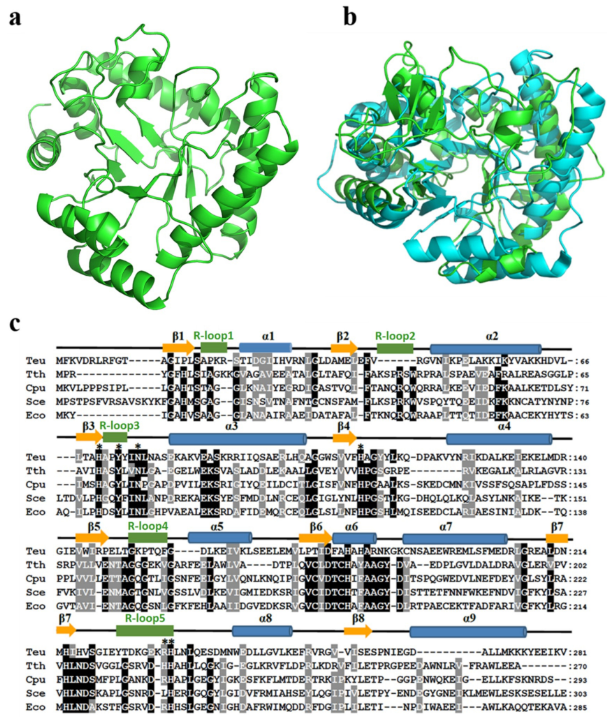
Homology model of TeuendoIV and multi-alignment of EndoIVs. (a) Modeled structure of TeuendoIV. (b) Superimposition of TeuendoIV (green) and EcoendoIV (cyan). (c) Multiple sequence alignment of EndoIVs. The EndoIVs are from T. eurythermalis, T. thermophilus (PDB ID: 3AAM), Chlamydophila pneumoniae, S. cerevisiae and E. coli (PDB ID: 1QTW). The secondary structures of EcoendoIV are shown at the top of sequences. Cylinders indicate α-helices, arrows indicate β-strands, and five green rectangles indicate DNA-binding recognition loops 1–5. Identical and similar residues are shaded in black and gray, respectively. The mutated residues are marked by asterisks.

