Unexplored diversity and ecological functions of transposable phages
Phages are prevalent in diverse environments and play major ecological roles attributed to their tremendous diversity and abundance. Among these viruses, transposable phages (TBPs) are exceptional in terms of their unique lifestyle, especially their replicative transposition. Although several TBPs have been isolated and the life cycle of the representative phage Mu has been extensively studied, the diversity distribution and ecological functions of TBPs on the global scale remain unknown. Here, by mining TBPs from enormous microbial genomes and viromes, we established a TBP genome dataset (TBPGD), that expands the number of accessible TBP genomes 384-fold. TBPs are prevalent in diverse biomes and show great genetic diversity. Based on taxonomic evaluations, we propose the categorization of TBPs into four viral groups, including 11 candidate subfamilies. TBPs infect multiple bacterial phyla, and seem to infect a wider range of hosts than non-TBPs. Diverse auxiliary metabolic genes (AMGs) are identified in the TBP genomes, and genes related to glycoside hydrolases and pyrimidine deoxyribonucleotide biosynthesis are highly enriched. Finally, the influences of TBPs on their hosts are experimentally examined by using the marine bacterium Shewanella psychrophila WP2 and its infecting transposable phage SP2. Collectively, our findings greatly expand the genetic diversity of TBPs, and comprehensively reveal their potential influences in various ecosystems.
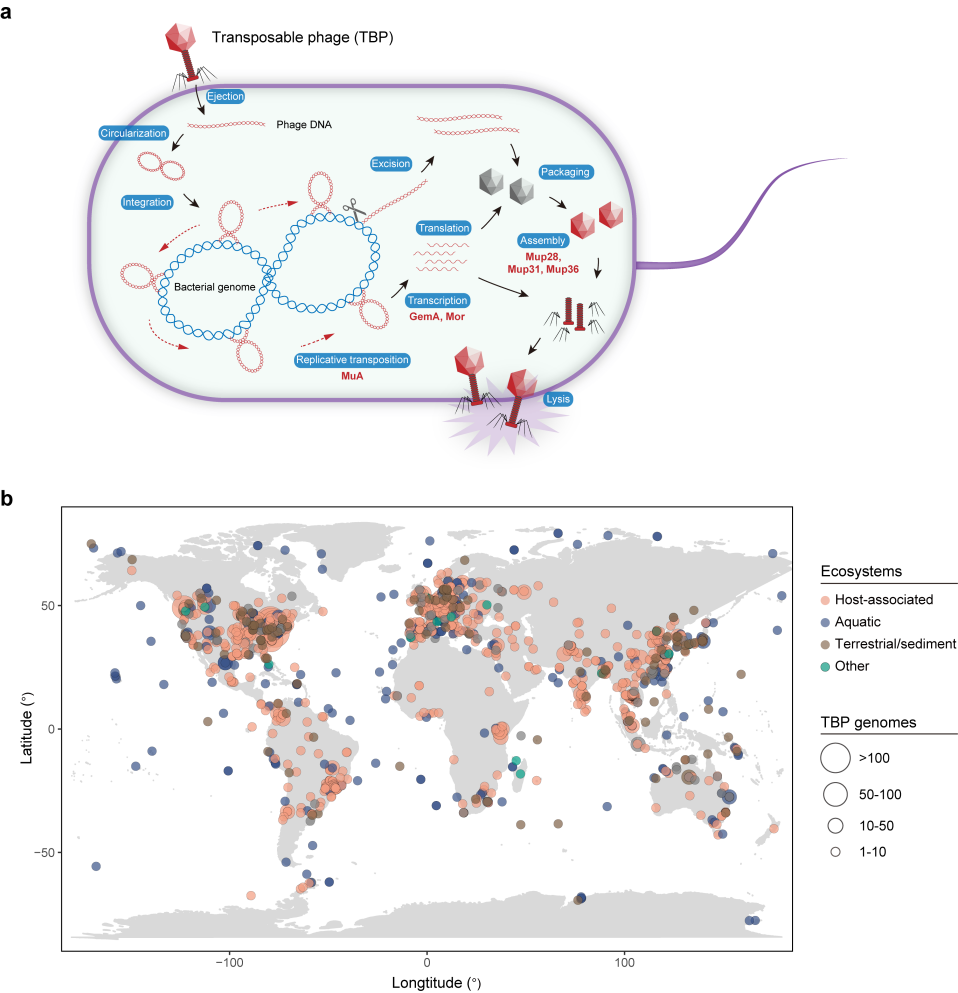
a Schematic life cycle of the TBPs. The six conserved marker proteins of TBPs are indicated in red. For clarity, the phage genomic DNA and virions are not to scale relative to the bacterial host cell and chromosome. b Geographic distribution of TBPs. Each point represents a geographic site, and only the TBP genomes (n = 5320) with available geographic coordinates are shown. The size of each point is proportional to the number of genomes found at that site, and the colors differentiate the derived environments: host-associated (orange), aquatic (blue), terrestrial/sediment (brown) or other (environmental information unavailable, green).
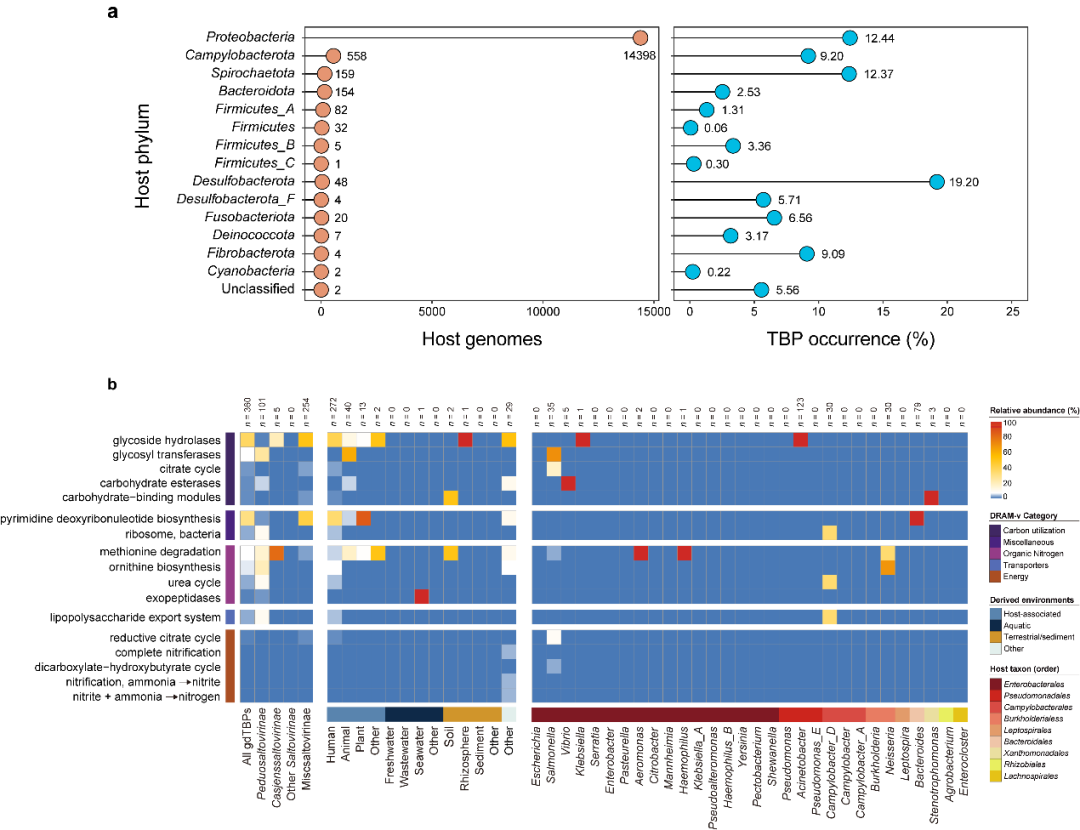
a Phylogenetic distribution of TBP hosts in different bacterial phyla. The values next to each circle represent the number of recovered TBP genomes (left panel) or TBP occurrence (right panel) for the specific bacterial phylum. b The heatmap shows the relative abundance and functional category of AMGs in each grouping. Only the AMG-encoding TBPs were included in the analysis. The number of TBP genomes contained in each group is shown at the top of the heatmap. The AMGs were identified and annotated by DRAM-v. MISC, miscellaneous.

a Genomic map of the prophage SP2 in the marine bacterium Shewanella psychrophila WP2. The arrows depict the location and direction of predicted proteins on the phage genomes, and the fill colors indicate different functional categories of genes, as indicated in the legend. b Graphic display of differentially expressed genes (DEGs) categorized by function in S. psychrophila WP2 after SP2 deletion. The transcriptome data represent three biologically independent samples for each strain (WP2 and WP2ΔSP2). Normalized differential expression levels (fold changes of WP2ΔSP2 versus WP2) are represented by heatmaps in boxes according to the scale bar (log2 scale) from most upregulated (red) to most downregulated (blue). The proteins encoded by the DEGs are shown in each box

