Widespread Bathyarchaeia encode a novel methyltransferase utilizing lignin‐derived aromatics
Lignin degradation is a major process in the global carbon cycle across both terrestrial and marine ecosystems. Bathyarchaeia, which are among the most abundant microorganisms in marine sediment, have been proposed to mediate anaerobic lignin degradation. However, the mechanism of bathyarchaeial lignin degradation remains unclear. Here, we report an enrichment culture of Bathyarchaeia, named Candidatus Baizosediminiarchaeum ligniniphilus DL1YTT001 (Ca. B. ligniniphilus), from coastal sediments that can grow with lignin as the sole organic carbon source under mesophilic anoxic conditions. Ca. B. ligniniphilus possesses and highly expresses novel methyltransferase 1 (MT1, mtgB) for transferring methoxyl groups from lignin monomers to cob(I)alamin. MtgBs have no homology with known microbial methyltransferases and are present only in bathyarchaeial lineages. Heterologous expression of the mtgB gene confirmed O-demethylation activity. The mtgB genes were identified in metagenomic data sets from a wide range of coastal sediments, and they were highly expressed in coastal sediments from the East China Sea. These findings suggest that Bathyarchaeia, capable of O-demethylation via their novel and specific methyltransferases, are ubiquitous in coastal sediments.
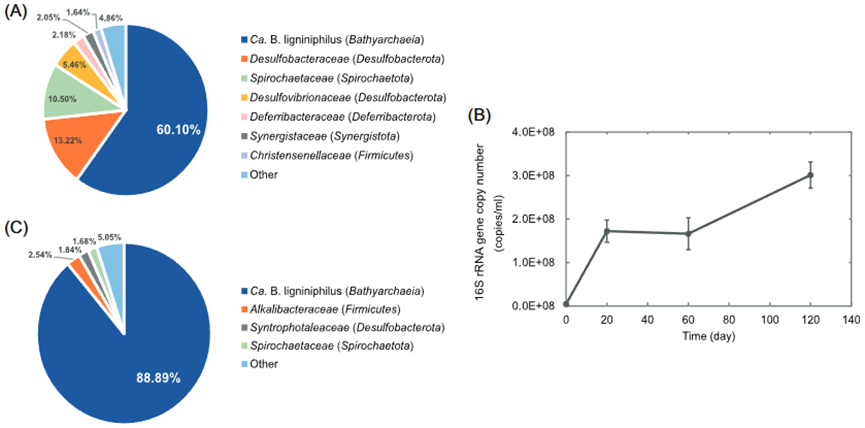
Figure 1. Microbial composition of enrichment cultures and growth curves of the cultured Ca. Baizosediminiarchaeum ligniniphilus. (A) Relative abundance of microbial populations based on the 16S rRNA gene-tag sequencing analysis before purification by adding antibiotics. (B) Growth curves of Ca. B. ligniniphilus in anaerobic medium supplemented with lignin. The error bars were obtained from triplicate qPCR reactions. (C) Relative abundance of microbial populations based on the 16S rRNA gene-tag sequencing analysis after purification by adding antibiotics.

Figure 2. An overview of the central metabolic pathway of Ca. Baizosediminiarchaeum ligniniphilus. The process of O-demethylation of guaiacol is accompanied by the fixation of carbon dioxide and the production of acetate. Yellow rectangles of varied shades show different gene transcription levels and blue rectangles of different shades show whether the relevant proteins were detected in the proteomics. Below the pathway is the Gibbs free energy (ΔG) estimated for the overall equation. FPKM, fragments per kilo base of transcript per million fragments mapped.
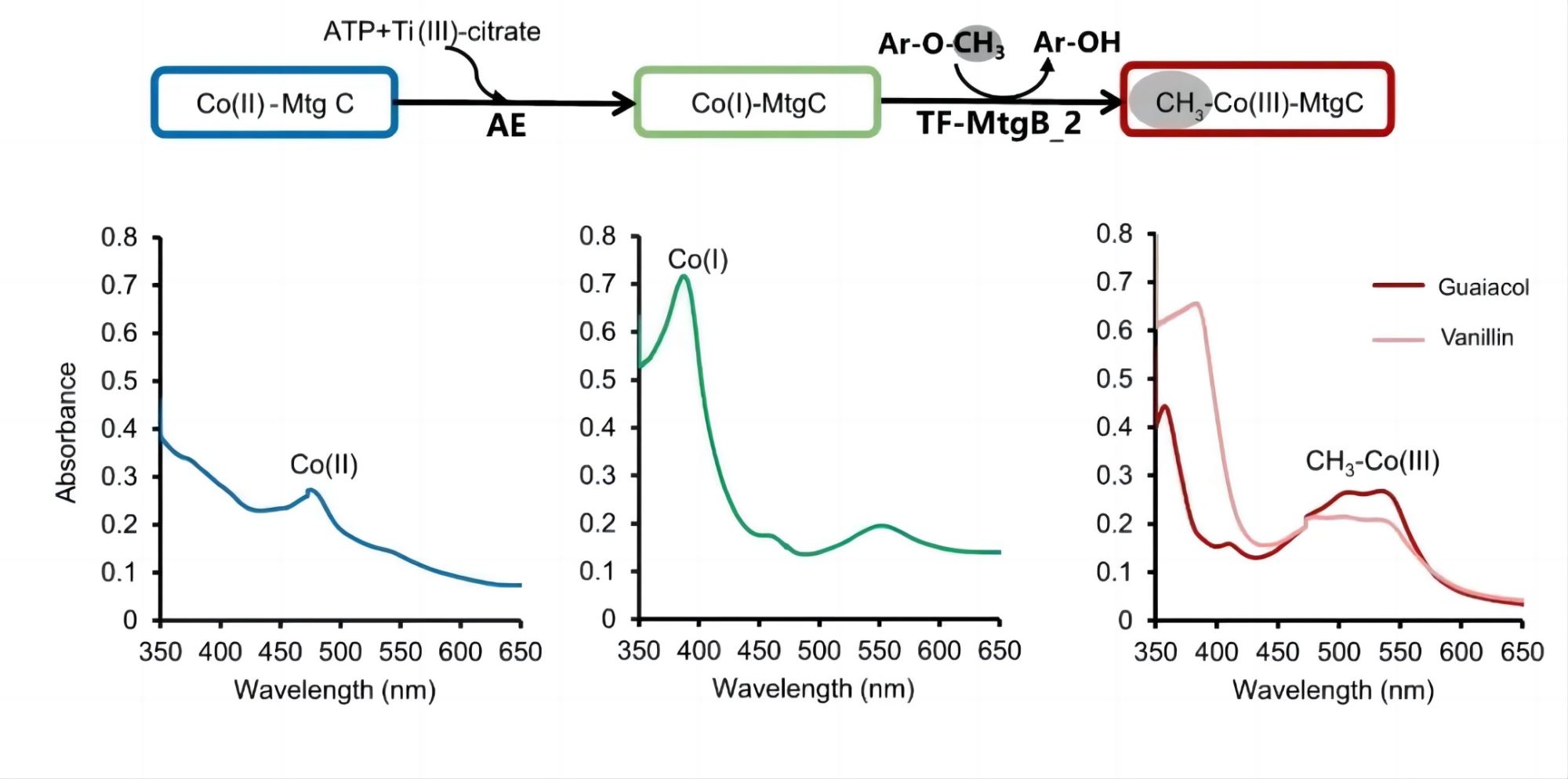
Figure 3. The O-demethylation/methyltransferase activities of Ca. B. ligniniphilus with guaiacol and vanillin as the substrates. Reaction 1: Activating enzyme (AE), ATP, and titanium (III) citrate are required for the activation of corrinoid protein (MtgC) from the Co(II) state (blue) to the active Co(I) state (green); AE was from Acetobacterium dehalogenans DSM 11527 (GenBank accession no. ACJ01666.1). Reaction 2: MtgB transfers the methyl group from guaiacol to Co(I)-MtgC, resulting in methylated Co(III)-MtgC (red and pink). All bottom panels correspond to UV/visible spectra measured after each reaction, reflecting the different states of the cobalamin carried by MtgC. Ti(III), titanium(III).
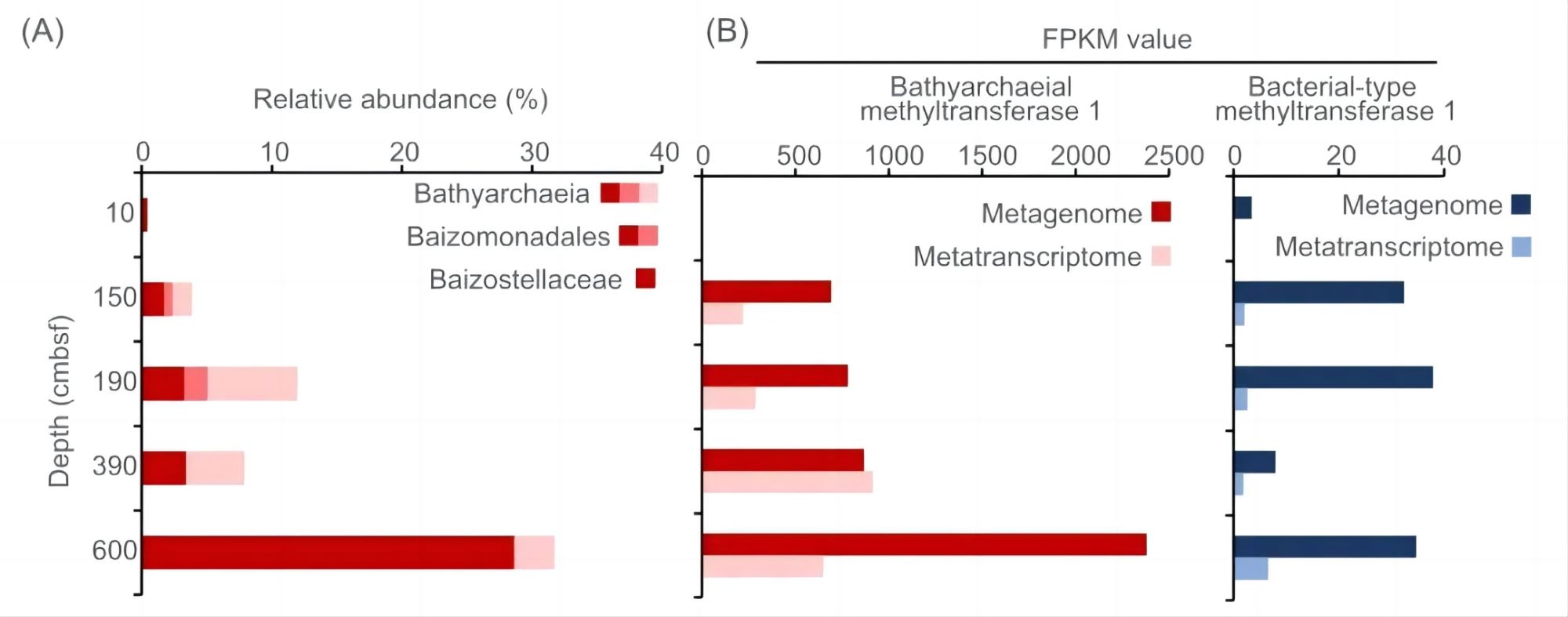
Figure 4. Distribution of the Bathyarchaeia-specific methyltransferase 1 (MT1, mtgB) genes in coastal sediments from the East China Sea. (A) The relative abundance of Bathyarchaeia. (B) The gene abundance and gene expression level of Bathyarchaeia-specific MT1 (red) and bacterial-type MT1 (blue).

